Enabling scientific research on the global biodiversity of food
Welcome to The Periodic Table of Food Initiative (PTFI)® Research Hub
We support scientific research and interventions through the application of multi-omics tools, enabling deep discovery and critical analysis of environmental, agricultural, and cultural factors, and illuminating the global impact on food quality based on diverse components and implications for human and planetary health.
Log in, or register your account to access PTFI tools and data.

The Periodic Table of Food Initiative is a global effort to provide data-driven solutions to transform food systems for improved human and planetary health.
The PTFI provides standardized tools, data, and training to map food quality of the world's edible biodiversity. We envision a world where each stakeholder involved in food and health systems is empowered to lead data-driven solutions for enhanced human and planetary wellbeing.
Learn more about the PTFI at foodperiodictable.org
PTFI Research
The PTFI's analytical approach is characterized by its collaborative, standardized, and distributable nature.
Collaborative
The PTFI takes a collaborative approach to standardizing Foodomics tools, engaging lab partners around the globe in the evaluation, development and validation of multi-omics protocols. These platforms currently include untargeted metabolomics, lipidomics, ionomics and fatty acid analysis. Additional platforms in development include glycomics, targeted metabolomics, proteomics and aromatics. Learn more
Standardized
The PTFI has four key areas of standardization to enable distributable analytical methods to labs around the world, including:
- Standardized protocols for each multi-omics platform, which will be publicly accessible at launch
- Custom internal standards to accompany the PTFI methods that allow for harmonization of data across labs, allowing the global ecosystem of PTFI partners to generate and contribute food composition data to the PTFI database
- An expanding cloud-based chemical library that allows for confident annotation of features detected in foods, without the need for labs to create their own chemical libraries
- A centralized data pipeline that allows labs to upload raw data from their instruments via the PTFI lab portal for data processing, retention time alignment, and final data assembly
Distributable
The PTFI’s overarching goal is to enable labs around the world to access methods, participate in food composition analysis, and contribute to building the largest database of food biomolecular composition to date.
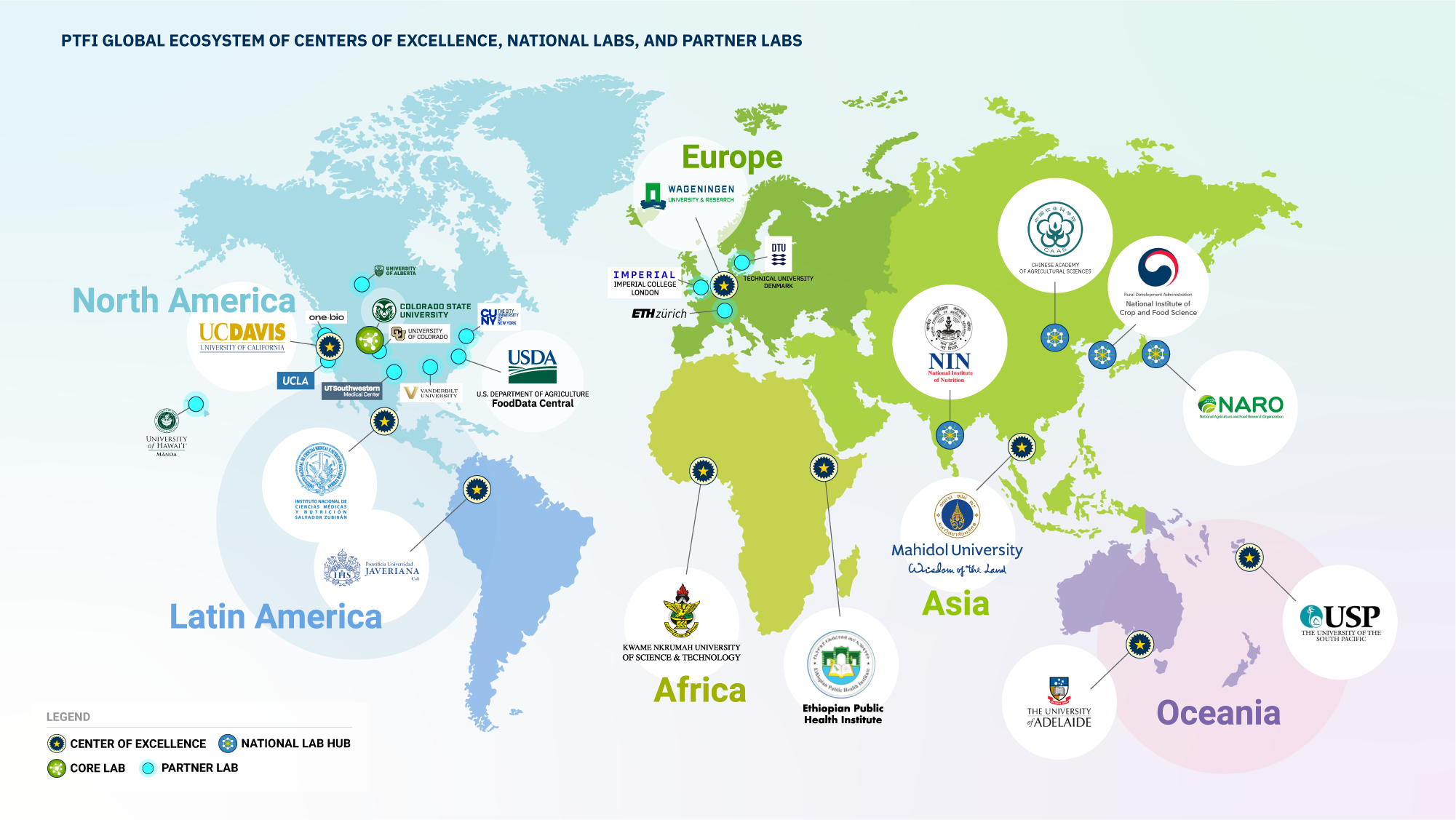
We’re currently working with nine Centers of Excellence around the globe to onboard PTFI methods and enable them to serve as PTFI research hubs in their geographic regions. The PTFI methods, reagents, chemical library, and data pipeline will also be available for individual labs to participate in this global effort.
PTFI Data
Standardized PTFI methods were utilized to characterize the biomolecular composition of 500 foods, together with their standard metadata, showcasing a wide array of diversity spanning 250 unique species and 56 unique food ontologies from Actinidiaceae to Zingiberaceae. These 56 ontologies can be categorized into six FoodOn categories, a globally harmonized food ontology system describing food from production to consumption. Among the 500 foods, there are 46 plants (including 10 types of vegetables and 37 types of fruits), 19 animal species (both domesticated and wild), algae, fungi, bacteria, and prepared meals. These foods were sourced from the United States and originate from 11 distinct countries, encompassing various production methods (e.g., organic vs. conventional) and preparation techniques (e.g., raw vs. cooked).
The multi-omics protocols used to generate the data include lipidomics, ionomics, glycomics, fatty acid analysis, proteomics, and untargeted metabolomics.
Tools & Resources

Multi-omics data organized and presented in an intuitive, engaging and enlightening format.
MarkerLab is a web-based data visualization platform that makes multi-omics data accessible, interpretable, and valuable for labs and researchers.
Explore PTFI Data with MarkerLab
The Periodic Table of Food Initiative (PTFI) is mapping food quality based on multi-omics data around the globe to improve human and planetary health. Researchers can use MarkerLab to search the PTFI database and explore food biomolecular data using a variety of visualization options. In addition to interactive data plots and tables for individual samples, MarkerLab also provides the option to compare foods and compounds of interest across the database.
Register your Research Hub account to access PTFI data with MarkerLab.


The American Heart Association’s Precision Medicine Platform is a cloud-based technology solution that allows researchers to collaborate and analyze large datasets from any computer in the world using a secure environment and the power of machine learning.
Within the research interface, users can request access to assorted datasets, including the Periodic Table of Food Initiative database to accelerate findings into impactful discoveries.
Researchers can analyze raw PTFI data to gain insights and run analyses utilizing pre-installed analytic tools. The AHA Data Science team provides a tutorial notebook in the workspace to guide users on how to view the dataset and run summary statistics. This tutorial is intended to help jumpstart analysis of the PTFI data on the Precision Medicine Platform.
Visit the Precision Medicine Platform’s PTFI data resource page for more information about requesting a workspace to analyze the PTFI data.
PTFI Research & Development Partners